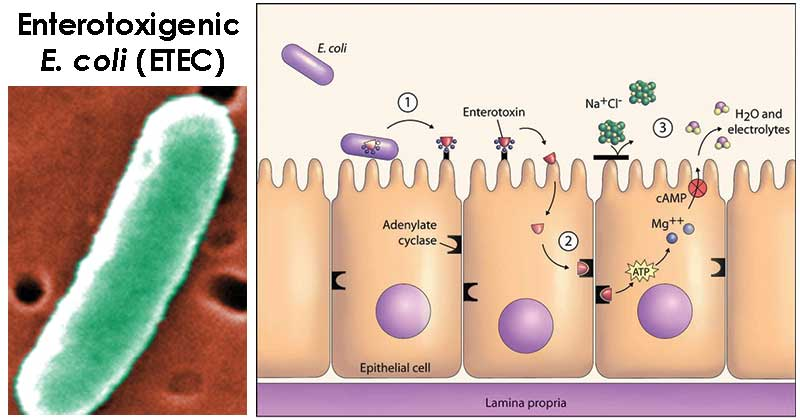
As much as we love to travel, nothing ruins a trip like a bout of diarrhea. While often dismissed as a minor inconvenience, it can be a serious problem in endemic areas, where it can cause severe dehydration and even death in young children. Enterotoxigenic Escherichia coli (ETEC) is one of the main culprits behind this scourge, causing millions of cases every year. But what if we told you that the solution to this problem might lie in the very molecule that helps digest our food?
This idea is the focus of Åsa Sjöling’s research at the Karolinska Institutet, where she studies the interaction between ETEC and bile salts. Bile salts are molecules produced by our liver and released into the gut after a meal to aid in digestion. While most bacteria can’t survive in the harsh environment of the gut, some pathogenic species, such as ETEC, have evolved to use specific bile salts as a signal to turn on virulence.
In previous work, Sjöling and her team found that one such bile salt, glycocholate, can upregulate the ability of ETEC to adhere to intestinal cells, making it more pathogenic. The current project, funded by the PAR Foundation, aims to build on these findings by studying the molecular mechanisms behind this process and identifying potential protective bacteria that can decrease the amount of glycocholate in the gut.
To do this, the team is using cutting-edge techniques such as organoids and deep metagenome sequencing. Organoids are three-dimensional structures that mimic the architecture and function of human organs, allowing researchers to study the interaction between ETEC and intestinal cells in a more realistic environment. However, adding bile salts to these cultures damages the cells, making this part of the project unfeasible. Instead, the team is focusing on determining the molecular adherence of glycocholate to a specific transcription factor, as well as identifying potentially protective bacterial species by co-culturing ETEC with an anaerobic gut bacteria culture and analyzing the shifts in prevalence using deep metagenome sequencing.
While it’s still early days, the results so far are promising. The team has identified minor shifts in the prevalence of certain Bacteroides and Clostridium species, which are normally present in the human gut, and plans to study the interactions between these species and ETEC in future experiments.
Subscribe to our Newsletter to stay updated on news from our grantees